This is blog about an experiment that feels like a personality test. 😅 So, somewhere between my third cup of coffee and the moment I realized I’d been wearing my lab coat inside-out for an entire morning, I decided today was going to be a 3’ RACE day. 🥲☕️
There’s a specific kind of confidence you get as a postdoc. Not the “I know everything” kind—more like the quiet, haunted confidence of someone who has already survived enough failed PCRs to develop a sixth sense for when a reaction is about to disappoint you.
So when I tell you I’m doing 3’ RACE, what I really mean is: I’m about to politely ask a transcript how it ends, and it’s going to make me work for it. 😭🧬
Because that’s what 3’ RACE is: A method for amplifying the sequence from a known internal region of an mRNA to its unknown 3’ end, by using the poly(A) tail as your universal anchor.
Scientifically, it’s beautiful.
Emotionally, it’s like texting someone “hey” and watching them type… then stop… then leave you on read. 📱💔
My Brain: “This should be simple.” (Narrator: it wasn’t.) 😌
3’ RACE looks straightforward on paper: make first-strand cDNA using an oligo(dT) adapter primer PCR using one gene-specific primer (GSP) + a universal primer nested PCR for better specificity gel → clone → sequence → finally know the 3’ end
🙂 In reality, 3’ RACE is never “just PCR.” It’s the molecular biology version of: “I know you exist… I just need proof.” 🕵️♀️🔍
Anyway. Let’s RACE.
Act I: RNA Extraction — The Part Where You Realize RNA is a Drama Queen 🧊🧤
Before we even get to the fun stuff, we have to talk about RNA quality, because 3’ RACE doesn’t tolerate bad RNA. The manual is very clear: RNA integrity affects how much full-length sequence you can capture.
And yes, I’ve learned to recognize the difference between: ✅ “RNA might be okay” and ❌ “RNA is absolutely not okay, but I’m going to try anyway because hope is a coping mechanism.” 😭😅😂
RNA degradation is the kind of pain that feels personal. And yes, contamination matters too. RT inhibitors like guanidinium salts, EDTA, SDS can mess up reverse transcription.
Which is why the protocol leans toward classic extraction methods (like guanidinium isothiocyanate + acid phenol) because they tend to produce cleaner RNA.
I know, I know…… If your RNA is trash, your day is trash. 🗑️🙂
Act II: The Poly(A) Tail — Nature’s Built-In Handle 🍑
The reason 3’ RACE works at all is because eukaryotic mRNAs usually have this elegant little feature at the end:
poly(A) tail = AAAAAAAA…
It’s like the transcript’s luggage handle. Or a wrist strap. 😂 Or that thing on suitcases that makes it easier to drag your entire life around. 🧳🧬
The method uses anchored PCR, meaning you amplify from a known internal sequence to the 3’ end.
So I use an adapter primer (AP), which is an oligo(dT) primer containing a defined adapter sequence. That adapter is crucial, because later you get to use a universal amplification primer that recognizes it. 😌✨
Act III: Reverse Transcription — Turning RNA into Something PCR Can’t Ruin (Yet) 🧬➡️📄
Okay. cDNA synthesis time. The manual tells you to heat RNA + adapter primer at 70°C for 10 minutes, then chill on ice.
Okay! This step is basically: “Unfold yourself, transcript. I’m trying to talk to you.” 😭
Because RNA secondary structures can block primer annealing. And RNA loves folding. It folds like it has hobbies.
Then you add SuperScript II RT and do: 42°C for ~50 min 70°C to terminate.
SuperScript II is engineered to have reduced RNase H activity, which helps preserve full-length cDNA.
Well, this is one of those moments where you appreciate enzyme design on a spiritual level. Because you’ve worked with enzymes that behave like: “I will copy your RNA, but I will also randomly ruin your day.” 😌🫠 SuperScript II is at least a little more emotionally stable.
Act IV: RNase H — The “Remove the Original” Step 🗑️
After RT, you add RNase H and incubate at 37°C to digest the RNA strand in the RNA:cDNA hybrid.
RNase H improves PCR sensitivity because it frees up your cDNA. This step always feels like a metaphor.
RNA: “I carried the message.”
RNase H: “Thank you. You may now perish.” 👋🙂
Act V: PCR — The Part Where You Trust One Primer With Your Entire Reputation 😭
Here’s the insane part of 3’ RACE: During RT, that adapter primer binds to all polyadenylated transcripts, so your cDNA pool contains basically everything. But when you PCR, you only have one gene-specific primer to provide specificity.
So you’re basically saying: “Dear PCR… please only amplify MY transcript.”
PCR: “lol” 😭
The PCR uses: GSP (gene-specific primer) AUAP or UAP (universal primer binding the adapter)
This is the entire “anchored PCR” strategy in action.
And yes, it’s elegant.
And yes, it’s fragile.
Like my work-life balance. 🥲⚖️
AUAP vs UAP: choose your toxicity level 💅
The kit I use offers two universal primer options: 👀AUAP: a clean universal primer that matches the adapter sequence. 👀And UAP: includes a dUMP-containing sequence for downstream UDG cloning.
But the UAP has drama: 🤔It shouldn’t be used with certain high-fidelity archaeal polymerases or long PCR mixes containing them because dUMP residues can inhibit those polymerases.
So AUAP is the “stable relationship,” and UAP is the “exciting but complicated” option. 😭💔
Act VI: Your Gel Looks Like Abstract Art (but you don’t have to care)🎨
The manual discusses free Mg²⁺ being important for PCR efficiency, and how Mg²⁺ interacts with dNTPs.
If your primer is not extremely specific, you may get multiple bands. The kit literally warns that multiple products can appear depending on GSP specificity.
And here’s the truth: Seeing a smear doesn’t just hurt because it failed. It hurts because you did everything right. You aliquoted the reagents. You used fresh tips. You whispered encouragement to your thermal cycler. You even held your breath during pipetting like it mattered. 😭🧪
And still your gel is like: “I made 9 products. None of them are yours.” 🫠
But the manual reminds us of the real goal: Eventually, 3’ RACE should give a single prominent band on the gel.
Keyword: eventually.
As a postdoc, “eventually” is basically my job description. 😭
Act VII: Nested PCR — When You Stop Asking Nicely 🔒
Nested PCR is where you get serious. You design a second gene-specific primer that binds inside the first PCR product, and run a second PCR. This dramatically improves specificity and gives you the effect of having another gene-specific priming event. Nested PCR feels like:
First PCR: “Maybe it’s my transcript?”
Second PCR: “No. I want certainty.” 😤✅
And when nested PCR works, it’s honestly one of the most healing feelings in molecular biology. Because suddenly, your chaos becomes a result.
Act VIII: Cloning & Sequencing — Proof That You Were Right All Along 📎
Once you’ve got your band, you can clone it. The kit I use even designs its primers with cloning in mind😳😳😳—adapter primer includes restriction sites and a NotI half-site to help cloning strategies.
UAP supports UDG cloning strategies and AUAP supports other cloning workflows; both can help generate directional clones depending on how you engineered your GSP tails.
And then sequencing finally gives you the truth: your real 3’ UTR your real polyadenylation site that sneaky alternative isoform the reason your qPCR primers have been acting suspicious 😭 And this is the part where you sit back and whisper:
I KNEW IT. 😌✨
The Ending: When the Band Shows Up, It’s Not Just a Band 🥹
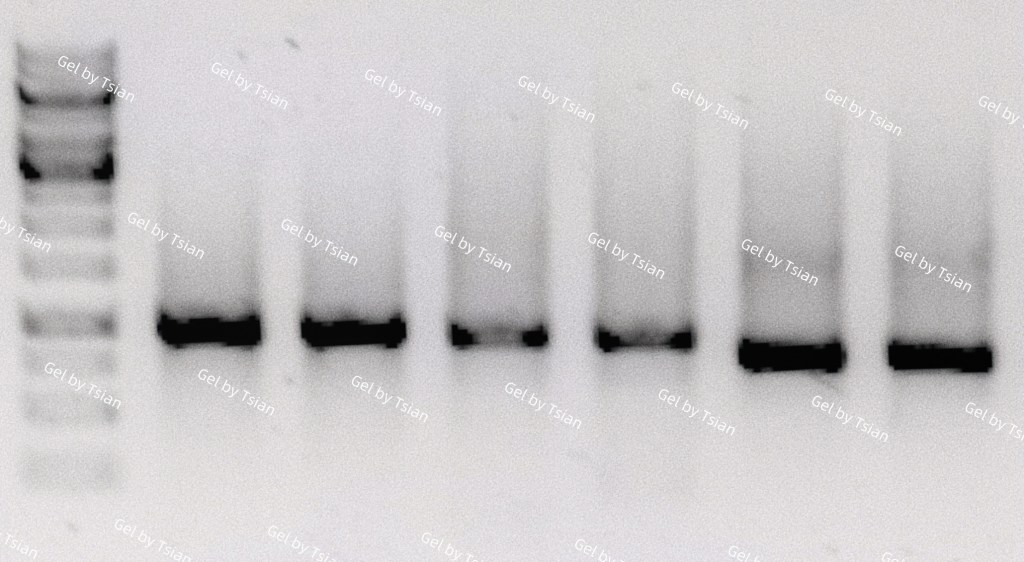
People who haven’t lived in a lab think a PCR band is “just DNA.”
But we know better.
That band is: your primer design skills your RNA integrity your ability to troubleshoot under psychological warfare your capacity to keep going even after 3 failed gels your stubbornness, disguised as “science”.😭❤️
Note: This whole adventure was done with the 3′ RACE System for Rapid Amplification of cDNA Ends (Invitrogen, Cat# 18373-019). The kit provided primers, enzymes, and instructions; I provided the overthinking, troubleshooting, and a suspicious number of agarose gels. 🧫😮💨
